Sequence database setup: UniProt proteomes
Overview
A UniProt complete proteome consists of the set of proteins thought to be expressed by an organism whose genome has been completely sequenced. A reference proteome is the complete proteome of a representative, well-studied model organism or an organism of interest for biomedical research.
UniProtKB is a collaboration between the European Bioinformatics Institute, the Swiss Institute of Bioinformatics and the Protein Information Resource.
Download
|
Fasta files representing the proteome for an organism can be downloaded by searching for a specific taxonomy accompanied by the keyword "Complete proteome":
- Perform the query and view the resulting list of entries (e.g. organism:9606 AND keyword:”Complete proteome” for the human proteome
- Click the orange Download button in the query result page
- Choose Fasta, Canonical and isoform sequence data in FASTA format
For example, to get the complete proteome for rice, search for taxonomy:4530 AND keyword:"Complete proteome".
In Database Manager, create a new custom definition using UniProt_proteome_template as the template.
You can enable automatic updating of a
UniProt Proteome by setting the Fasta file URL. Just change the taxonomy ID in this sample URL to
the one for your proteome of interest:
http://www.uniprot.org/uniprot/?query=taxonomy:4530+AND+keyword:"Complete+proteome"&force=yes&format=fasta&include=yes
The complete configuration for the rice proteome in Database Manager will look similar to this

Taxonomy
Taxonomy is not required for a single organism database
Parse Rules
When a single entry is expanded into entries for multiple isoforms, they share the same ID, so AC must be used as the unique identifier
>sp|Q67W82-2|4CL4_ORYSJ Isoform 2 of Probable 4-coumarate--CoA ligase 4 OS=Oryza sativa subsp. japonica GN=4CL4
AC from Fasta title: ">..|\([^|]*\)"
Description from Fasta title: ">[^ ]* \(.*\)"
Configuration (Mascot 2.3 and earlier)
A Fasta file containing canonical and isoform sequence for the rice proteome was downloaded to /usr/local/mascot/sequence/rice_proteome/current, and renamed to rice_proteome_20120414.fasta.
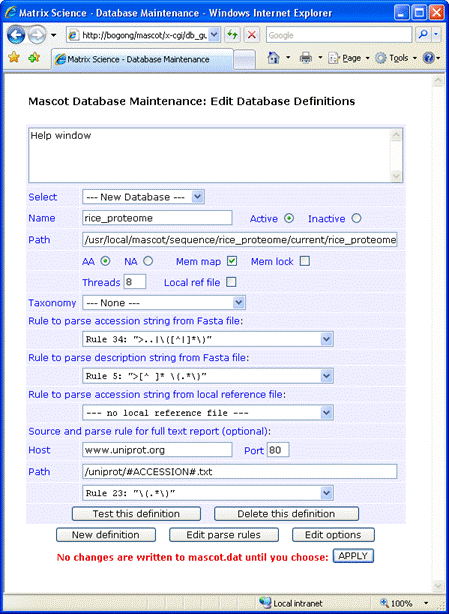
Full text for individual entries can be retrieved across the web from Uniprot:
Host: www.uniprot.org
Port: 80
Path: /uniprot/#ACCESSION#.txt
Parse rule: RULE_23 "\(.*\)"
Always test a new definition before applying the changes to mascot.dat